Genetic basis for tissue-specific DNA methylation patterns in plants
How does a cell develop into various types of tissues and organs with the same set of genome? Epigenetic modification patterns play a vital role in these processing. DNA methylation, a type of stably inheritable epigenetic modification, which is ubiquitous and conserved in both animals and plants, are essential for gene regulation, transposon silencing and gene imprinting. However, how DNA methylation patterns are regulated has remained obscure in the field of epigenetics.
Recently, Assistant Prof. ZHOU Ming from the Zhejiang University College of Life Sciences and Associate Prof. Julie Law from the Salk Institute for Biological Studies in the United States co-published a research article entitled “The CLASSY family controls tissue-specific DNA methylation patterns in Arabidopsis” in the journal Nature Communications. They addressed the basic question that how DNA methylation patterns are regulated in different types of tissues using the model plant, Arabidopsis.
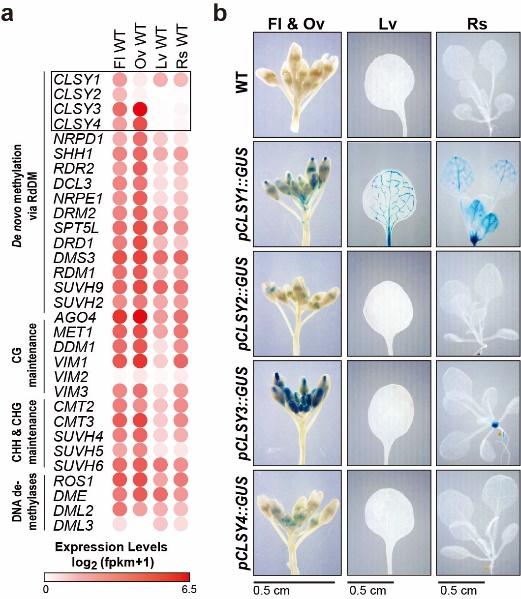
Fig.1 Tissue-specific expression patterns of the four CLSY genes
In this study, researchers first revealed the striking tissue-specific expression patterns of the CLSY genes in Arabidopsis. They found that the four CLSYs showed unique expression patterns in different tissues: CLYS1 and CLYS3 were highly expressed in the vascular tissues and ovules, respectively, whereas CLYS2 and CLYS4 expressed at low levels (Fig.1), suggesting CLSYs might play important roles in shaping epigenetic landscapes in plant.
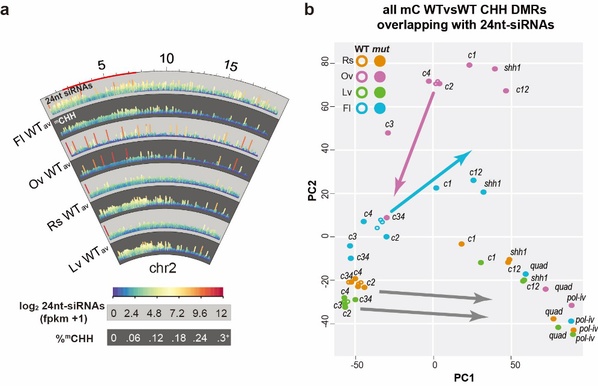
Fig.2 Impacts of the four CLSYs on genome-wide epigenetic landscapes
Integration of genome-wide small RNA-seq (smRNA-seq), MethylC-seq, mRNAseq and ChIP-seq in different combination of clsy mutants revealed that CLSYs shaped the plant epigenetic landscapes in a tissue-specific manner, and even a single clsy mutant (e.g. clsy3) was efficient to shift the epigenetic landscape between tissues (Fig.2). This work reveals how diverse patterns of methylation are generated during plant development.
This work expands previous findings that CLSYs act as locus-specific regulators for establishment of DNA methylation to define where to methylated (Zhou et al., Nature Genetics, 2018) and the current work determines which tissue for methylation. Together, these findings shed light into the possibility in altering DNA methylation patterns in much higher precision--in a tissue-specific or cell-type specific manner. Given the conservation of the DNA methylation systems in plants and mammals, these findings have broad implications for yield improvement for crops and precise gene therapy for human.