ZJU scientists reveal transcription activation by a sliding clamp
For decades, transcription of bacteriophage T4 late genes has served as a model system to investigate mechanisms of transcription regulation. The T4 late promoters consist of a -10-like element placed at the position corresponding to the bacterial promoter -10 element. Nevertheless, there is no sequence conservation at the position corresponding to the bacterial promoter -35 element. In recent years, the research team led by Prof. FENG Yu from the Zhejiang University School of Medicine has conducted a systematic research into the transcription activation of bacteriophage T4 late genes at a molecular level.
On February 18, the research team published an open-access article entitled “Transcription activation by a sliding clamp” in the journal Nature Communications. Transcription activation of bacteriophage T4 late genes is accomplished by a transcription activation complex containing RNA polymerase (RNAP), the promoter specificity factor gp55, the co-activator gp33, and a universal component of cellular DNA replication, the sliding clamp gp45. Although genetic and biochemical studies have elucidated many aspects of T4 late gene transcription, no precise structure of the transcription machinery in the process is available. It is the first time that researchers have reported the cryo-EM structures of a gp55-dependent RNAP-promoter open complex and an intact gp45-dependent transcription activation complex.
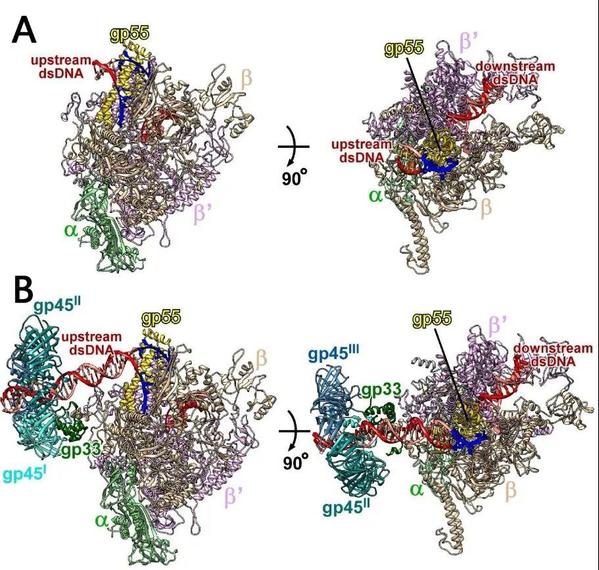
Gp55, a highly diverged σ70 family protein, plays an essential role in T4 late transcription by contacting the clamp helices and mediating the sequence-specific interactions with the promoter -10-like element. The structure of TAC-gp45 reveals that gp33 contacts the upstream dsDNA, which is crucial for gp45-depenent transcription activation. It also verifies that gp45 is located at the upstream end of TAC-gp45, in the vicinity of gp33. A model of gp45-dependent transcription activation has been proposed on the strength of previous studies. First, Gp45 is mounted onto DNA by gp44/gp62 complex. Then, RNAP is tethered to gp45 through gp55 and gp33. The quaternary complex scans for promoters along the DNA. If a T4 late promoter is encountered, the quaternary complex unwinds the promoter DNA and starts RNA synthesis. During transcription elongation, gp55 is displaced by the nascent RNA, but remains tethered to gp45. After transcription termination, the quaternary complex resumes promoter scanning without dissociation from the DNA. In this way, gp45 switches the promoter search from a three-dimensional diffusion mode to a one-dimensional scanning mode, thus tremendously boosting the efficiency of transcription activation.