Researchers discover a novel approach to labeling and imaging protein-coding genes in living cells
The research team headed by CHEN Baohui from Zhejiang University School of Medicine published an article entitled “Efficient labeling and imaging of protein-coding genes in living cells using CRISPR-Ta” in the November 29 issue of Nature Communications. Their studies open up a new technology for labeling non-repetitive genomic elements and display an efficient, easy, and scalable DNA tagging system in living human cells, thus laying a solid foundation for exploring the connection between gene positioning and gene expression.
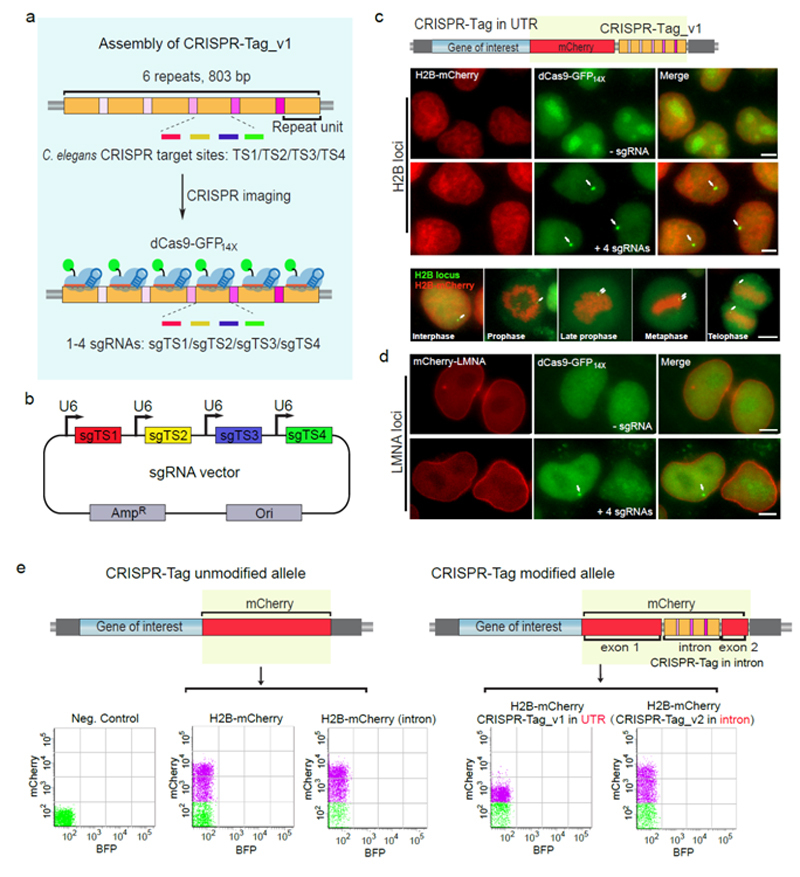
Individual genes and genomic regions are located at different positions in the three-dimensional space of the nucleus. Anomalies of the three-dimensional space are intimately bound up with many human diseases. The three-dimensional structure of genomes is marked by dynamism and heterogeneity of single-cells. Therefore, it is of supreme importance to investigate the spatiotemporal dynamics of such genes using a single-cell resolution.
Long-standing questions include whether the position of a gene affects its activity and how gene positioning is maintained and regulated. There is no doubt that utilizing imaging techniques, which allow the direct visualization of gene positioning and gene expression in living cells simultaneously, researchers are able to uncover how gene positioning is pertinent to gene activity. Recent efforts toward this end have focused on engineering a series of modular proteins with specific DNA recognition, including the clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated (Cas) system. The catalytically dead version of Cas9 (dCas9) has been extensively explored for imaging endogenous genomic loci in living cells. However, most of targets visualized by dCas9 system are still limited to the repetitive genomic region.
The major challenge is, when targeting non-repetitive genomic regions, it is essential that multiple sgRNAs function simultaneously to provide a sufficient signal-to-noise ratio for microscopy detection. For example, to visualize a non-repetitive gene or regulatory element in mouse embryonic stem cells, at least 26 sgRNAs need to be expressed from three CARGO arrays to achieve efficient labeling. Although two groups have reported that the number of sgRNAs could be reduced to 3–4 using a combination of signal amplification and super-resolution microscopy, the labeling efficacy has not been quantitatively assessed.
Well-designed approaches using CRISPR imaging as readouts are critical to further optimizing the DNA labeling system. CHEN Baohui et al. address this issue by developing a CRISPR-Tag system using one to four highly active sgRNAs to specifically label protein-coding genes with a high signal-to-noise ratio for visualization by wide-field fluorescence microscopy. This approach involves assembling a CRISPR-Tag within the intron region of a fluorescent protein and then integrating this cassette to N- or C-terminus of a specific gene, which renders possible simultaneous real-time imaging of protein and DNA of human protein-coding genes, such as HIST2H2BE, LMNA and HSPA8 in living cells.
This CRISPR-Tag system, with a minimal size of ~250 bp DNA tag, represents an easily and broadly applicable technique to study the spatiotemporal organization of genomic elements in living cells. Research into the relationship between gene positioning and gene activity will facilitate our perspicacious understanding of the three-dimensional structure of genomes.