Microwell-seq 2.0 for cost-effective and high-throughput screening with single-cell transcriptional profiling
The research team led by Prof. GUO Guoji at the Center for Stem Cell and Regenerative Medicine, the Zhejiang University School of Basic Medical Sciences, published a research article entitled “High-throughput Microwell-seq 2.0 profiles massively multiplexed chemical perturbation” in the journal Cell Discovery, bringing into public attention Microwell-seq 2.0—a high-throughput screening platform with single-cell multiomic profiling.
In 2018, GUO Guoji et al. created the first comprehensive mammalian cell map (the Mouse Cell Atlas) using Microwell-seq, a convenient, low-cost and robust platform for high-throughput single-cell RNA sequencing. This study was published in the journal Cell (Han X, Wang R, et al. 2018). To meet the growing demand for single-cell RNA sequencing, GUO Guoji et al. integrated pre-indexing technology (Rosenberg A, Roco C, et al. 2018; Cao J, Spielmann M, et al. 2019) with Mircowell-seq and established Microwell-seq 2.0 for the single-cell sequencing platform for high-throughput screening.
By using labeled reverse transcription (RT) primers and Tn5 transposase, researchers could pre-index the transcriptome and genome of a large number of cells. The labeled cells were then mixed and captured with an agarose plate, with each microwell able to trap multiple cells with different pre-labels. Cells in each microwell were labeled again through barcoded magnetic beads. In contrast with Microwell-seq 1.0, Microwell-seq 2.0 made better use of microwells. With two rounds of labeling, an agarose plate of Microwell-seq 2.0 could increase the throughput from 10,000 to nearly 1 million individual cells per experiment. Meanwhile, Microwell-seq 2.0 also gained an edge over the previous edition in dual-cell contamination control, sensitivity and cost control.
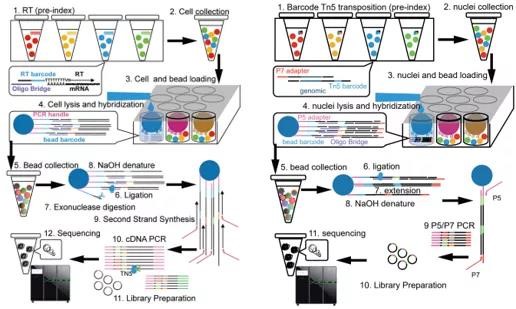
Schematic diagram of Microwell-seq 2.0 transcriptome (left) and ATAC (right) analysis
Cell-based high-throughput screening (HTS) is extensively applied in the field of life sciences. Assays suitable for HTS should be sensitive, robust, and economical. However, the readout of conventional HTS assays is restricted to gross phenotypes, including bulk transcriptional profiles, fluorescence signals, morphology, and viability, which cannot reveal subtle and heterogeneous changes in individual cells. In recent years, high-throughput single-cell sequencing technology has shown considerable promise in overcoming these limitations in cell-based HTS (Srivatsan, McFaline-Figueroa et al. 2020).
Prof. Guo led his team to analyze the perturbation of 48 small-molecule combinations on human embryonic stem cells using Microwell-seq 2.0, which exhibited exceptional sensitivity and robustness. Both Repsox and SB431542 are ALK inhibitors (Repsox: ALK5, ALK4, ALK7; SB431542: ALK5, TGFβR1). Microwell-seq 2.0 precisely identified their different perturbation effects.
Moreover, some small molecules, such as CHIR-99021, could significantly affect gene expression alone. However, other small molecules, such as retinoic acid, could produce apparent perturbations only when combined with other molecules.
“Our study illustrates the high sensitivity and robustness of Microwell-seq 2.0 in cell-based screening. This method may well pave the way for a more cost-effective multi-dimensional and high-throughput drug screening assay,” said Prof. Guo.