ZJU researchers crack the mystery of salicylic acid biosynthesis
In a major breakthrough that rewrites a classic paradigm in plant biology, scientists from Zhejiang University have uncovered the long-sought biosynthetic pathway of salicylic acid (SA) in plants — a discovery published in Nature on July 23, 2025. The study provides definitive molecular insight into how plants produce this critical hormone for immune defense, paving the way for engineering broad-spectrum disease resistance in crops.

SA, first identified in willow bark, is not only fundamental to the pharmaceutical development of aspirin, but also plays a central role in plant immunity. Despite its biological significance, the complete mechanism of SA biosynthesis in plants has remained enigmatic for decades.
Led by Dr. PAN Ronghui (Crop Science) and Dr. FAN Pengxiang (Horticulture) at Zhejiang University’s College of Agriculture and Biotechnology, the research team identified a new biosynthetic route involving three key enzymes that operate across different subcellular compartments. This newly characterized pathway redefines our understanding of SA production and provides a molecular “roadmap” for crop improvement.
The study’s co–first authors are Dr. WANG Yukang (postdoctoral fellow at the College of Agriculture and Biotechnology), Dr. SONG Shuyan (a researcher at ZJU-Hangzhou Global Scientific and Technological Innovation Center), and ZHANG Wenxuan (a doctoral student at the College of Agriculture).
“The prevailing theory held that benzoic acid is directly hydroxylated to produce salicylic acid, but the key enzyme for this step had never been found,” said PAN. “This knowledge gap has hindered our ability to design crops with enhanced disease resistance.”
Drawing on years of work on plant metabolic networks and subcellular biochemistry, the researchers initially identified a gene, CNL, which modulates the upstream pathway of SA biosynthesis. Using CNL as a molecular entry point and assisted by custom-developed and other published algorithms, the team performed large-scale multi-omics integration and discovered three essential enzymes — an acyltransferase, a hydroxylase, and a hydrolase — distributed in the peroxisome, endoplasmic reticulum (ER), and cytoplasm of rice cells.
Genetic knockout experiments confirmed the functional significance of these enzymes. “When we disrupted any of the three enzymes, SA production stalled,” said PAN. “This showed that a tightly coordinated enzymatic cascade drives SA biosynthesis in vivo.”
Surprisingly, these mutants failed to convert benzoic acid to SA, challenging long-held assumptions. “We repeatedly saw no increase in SA after feeding benzoic acid to the acyltransferase-deficient mutants,” said Wang. “That pushed us to think out of the box: benzoic acid might need to be converted into another compound first.”
When the researchers applied benzyl benzoate — a predicted intermediate — to the mutants, SA biosynthesis was restored. Isotope tracing then confirmed that benzoic acid was first transformed into benzyl benzoate. “That’s when it became clear: the canonical model was flawed. The true precursor isn’t benzoic acid, but benzyl benzoate,” said Zhang.
A major technical hurdle was confirming the hydroxylase activity in the ER, where it is uniquely active. “Conventional purification approaches failed,” said SONG. “Eventually, we detected its function by analyzing native membrane fractions from leaf tissue, and isotope labeling confirmed it catalyzed the conversion of benzyl benzoate into benzyl salicylate.”
Once these two major bottlenecks were resolved, the final hydrolysis reaction proceeded as predicted, completing the new three-enzyme pathway.
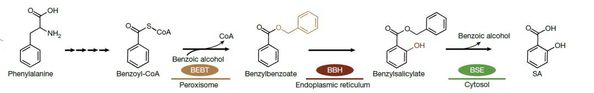
“This work definitively shows that benzoic acid must first be converted into benzyl benzoate to enter the salicylic acid pathway,” said FAN. “The discovery overturns a textbook model and offers precise molecular targets for next-generation crop breeding.”
The new biosynthetic route was validated across diverse crop species, including rice, tomato, wheat, and cotton. Through collaboration among multiple research groups in the university’s departments of Crop Science, Horticulture, and Plant Protection, the team demonstrated the broad conservation and functional relevance of this pathway across plant taxa.
“We now know this mechanism plays a crucial role in disease resistance across a wide range of important crops,” said PAN. “This kind of breakthrough was only possible thanks to deep interdisciplinary collaboration.”

CAE Fellow YU Jingquan, a member of the research team and a witness to the entire project, remarked: “This study is like ‘molecular archaeology’, decoding the biosynthesis of an ancient medicinal compound and a key plant hormone. More importantly, it equips us with new theoretical and technical tools to tackle global challenges in crop disease management, laying a scientific foundation for sustainable agriculture.”
Adapted and translated from the article by ZHA Meng
Translator: FANG Fumin
Photos by ZHE Yin
Editor: HE Jiawen, ZHU Ziyu