Evolutionary rate correlation discovered between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects
The research team led by Prof. YE Gongyin from the Institute of Insect Sciences, Zhejiang University, cooperated with Prof. John H. Werren from the University of Rochester to engage in research into the evolutionary rate correlation between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects. Their findings are published in the May 5 issue of the journal of Molecular Biology and Evolution.
The mitochondrion a pivotal organelle for energy production, and includes components encoded by both the mitochondrial and nuclear genomes. For example, the oxidative phosphorylation pathway (OXPHOS or OP) contains both mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins. Ribosomal RNAs (rRNA) are encoded by mitochondrial ribosomal RNAs (mrRNA) while nuclear-encoded mitochondrial ribosomal proteins (nMRP) are nuclear-encoded. It is only through the interaction between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins that mitochondria can function properly.
Then, how do nuclear- and mitochondrial-encoded components interact with each other in function and evolution? This topic is of immense interest in biology, with theoretical implications to genetics, evolution, and medicine. For example, a perspicacious understanding of the interaction between nuclear- and mitochondrial-encoded components is highly instrumental to exploring effective mitochondria replacement therapy. By the same token, in the field of pest control, the discovery of mitochondrial proteins with pest idiosyncrasies may well contribute to the screening of effective pesticide.
YE Gongyin et al. compare the evolutionary rates of mitochondrial proteins and ribosomal RNAs to rates of mitochondria-associated nuclear-encoded proteins, across the major orders of holometabolous insects. There are significant evolutionary rate correlations (ERCs) between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins, which are likely driven by different rates of mitochondrial sequence evolution and correlated changes in the interacting nuclear-encoded proteins. The pattern remains after correction for phylogenetic relationships and considering protein conservation levels. Correlations are stronger for both nuclear-encoded OXPHOS proteins that are in contact with mitochondrial OXPHOS proteins and for nuclear-encoded mitochondrial ribosomal amino acids directly contacting the mitochondrial rRNAs.
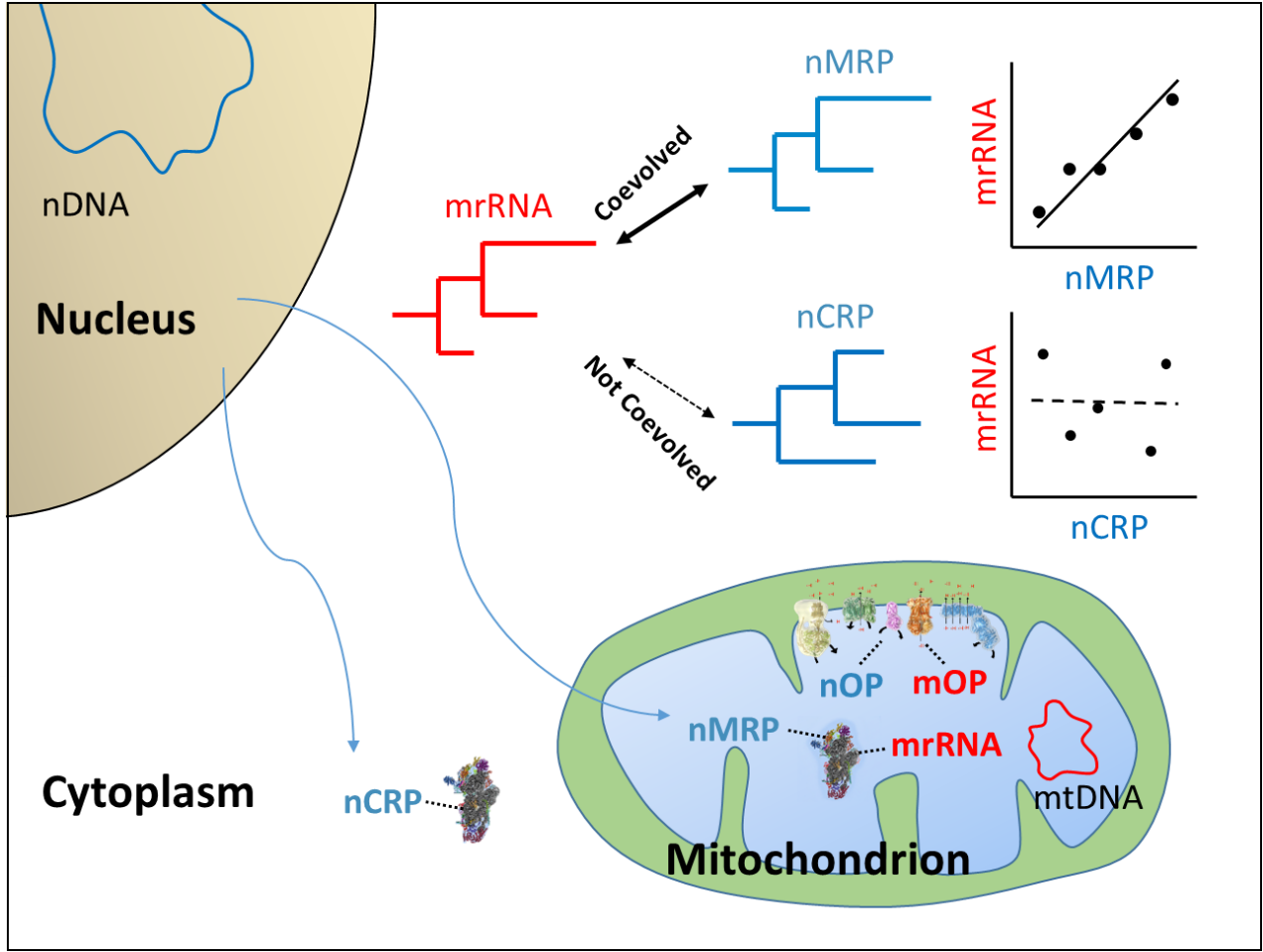
Researchers find that the ERC between mitochondrial- and nuclear-encoded proteins is a strong predictor of nuclear-encoded proteins known to interact with mitochondria, and it shows great promise for identifying new candidate proteins with mitochondrial function. Twenty-three additional candidate nuclear-encoded proteins warrant further study for mitochondrial function based on this approach, including proteins in the minichromosome maintenance helicase complex.





